A new study by Clare et al. has made significant strides in understanding the genetic basis of disease susceptibility in hybrid barley. A collaboration between researchers in the USA and China, co-ordinated by Washington State University, has identified and mapped a novel hybrid susceptibility locus, named Susceptibility to Pyrenophora teres 2 (Spt2), which is exploited by the necrotrophic pathogen Pyrenophora teres f. maculata (Ptm). This pathogen causes spot form net blotch (SFNB), a disease that can lead to severe yield losses. The innovative use of PACE® (PCR Allele Competitive Extension) SNP genotyping technology was critical to the success of this high-resolution mapping effort.
Significance to Barley Breeders
Hybrid barley, known for its potential yield advantages due to heterosis, can sometimes exhibit increased susceptibility to diseases. This phenomenon was observed in crosses between resistant barley lines (CI5791, Tifang, and Golden Promise) where the F2 progeny displayed extreme susceptibility to Ptm. Understanding the genetic underpinnings of this susceptibility is crucial for barley breeders aiming to develop resistant hybrid varieties. The discovery of the Spt2 locus offers valuable insights that can guide breeding strategies to avoid the release of hybrids with deleterious disease susceptibility traits.
Dr Shaun Clare, Lab Manager of the Barley Breeding and Molecular Genetics Lab at Washington State University, explains further: “Since the majority of barley production is not as a hybrid crop, this may seem like insignificant research. However, since we believe the molecular mechanisms underlying this hybrid susceptibility will also be relevant to the molecular control of hybrid vigor, we believe this will serve as an excellent model to really tease apart these complex genetic interactions”
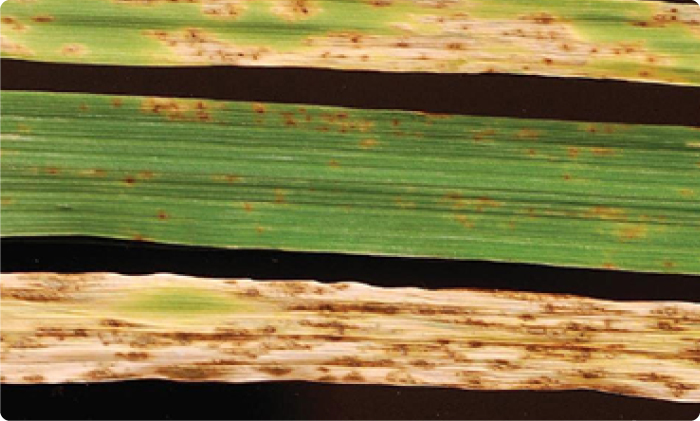
Phenotypic results of parental and hybrid barley. Parents C15791 (top), and Tifang (middle) and C15791 x Tifang F1 after one week post inculation with Pyrenophora teres f. maculata isolate 13IM8.3. From Clare et al. 2024.
PACE® Genotyping Technology
PACE genotyping played a key role in this research. It enabled the identification and mapping of genetic polymorphisms between different barley lines. By targeting specific polymorphisms, the researchers could precisely map the Spt2 locus to a 198 kb region on chromosome 5H. This high level of resolution was essential for identifying the candidate gene within the Spt2 locus, predicted to encode a pentatricopeptide repeat-containing protein (PPR).
Dr Clare continues, “Despite the rapid improvement in accuracy and cost of sequencing, sequencing all of our critical recombinants still would not be feasible. Converting polymorphisms identified through genomic comparison of the parental lines to PACE genotyping assays allowed us to really narrow down the final recombination points and delimit this locus to a single candidate gene”
Research Findings
The study involved comprehensive genetic mapping using F2 populations from CI5791 × Tifang and CI5791 × Golden Promise crosses. The susceptible phenotype segregated in a 1:1 ratio of resistant to susceptible individuals, indicating a single hybrid susceptibility locus. The mapping of Spt2 was independently validated in two different populations, strengthening the confidence in the results. Comparative genomic analysis between CI5791 and Golden Promise further supported the identification of a single candidate gene within the Spt2 locus.

Implications and Future Directions
The identification of the Spt2 locus and its candidate gene opens new avenues for research and breeding. Understanding how this gene contributes to hybrid susceptibility can help in developing strategies to mitigate this risk. Additionally, this study underscores the importance of considering hybrid susceptibility in breeding programs, especially for crops like barley, where hybrid varieties are increasingly being adopted.
Future research will focus on characterizing the Spt2 gene and its function in the context of hybrid susceptibility. Reverse genetic approaches, such as loss-of-function and knockdown studies, will be essential to validate the role of the PPR gene in disease susceptibility. Furthermore, exploring the genetic and epigenetic mechanisms underlying this susceptibility could provide deeper insights into how heterosis and hybrid vigor can be optimized without compromising disease resistance.
Conclusion
The work by Clare et al. highlights the innovative use of PACE genotyping technology for SNP genotyping to achieve high-resolution mapping of a novel hybrid susceptibility locus in barley. The study not only advances our understanding of hybrid susceptibility but also sets the stage for future research aimed at enhancing the resilience of barley crops to disease.


