Introduction: Understanding PCR Genotyping
PCR genotyping – also known as genotyping by PCR – is a powerful molecular technique used to identify genetic variation within DNA sequences. Leveraging the high specificity of polymerase chain reaction (PCR), researchers and breeders can accurately detect single nucleotide polymorphisms (SNPs) and insertions or deletions (Indels) with high accuracy. PCR genotyping offers a cost-effective, high-throughput, and scalable solution for SNP validation, marker-assisted selection (MAS), and genomic selection (GS) in both plant and animal breeding programs.

What is PCR Genotyping?
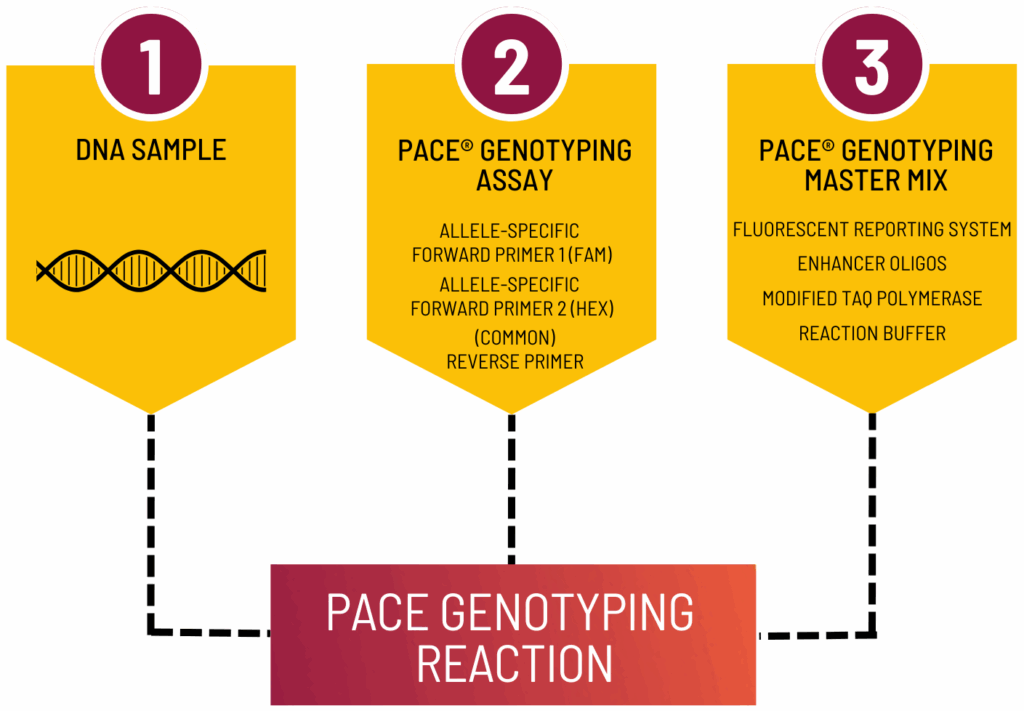
PCR genotyping involves amplifying specific DNA sequences using allele-specific primers to distinguish between genetic variants. Techniques like allele-specific PCR – including 3CR Bioscience’s proprietary PACE® chemistry– are widely adopted due to their accuracy, sensitivity, and adaptability to high-throughput formats. Unlike sequencing methods that require complex data processing, PCR genotyping delivers rapid, actionable insights into genetic traits with minimal sample preparation and runtime.
As von Maydell (2023) notes in “PCR allele competitive extension (PACE).” Plant Genotyping: Methods and Protocols. New York, NY: Springer US, SNPs—codominant, genome-wide, and highly abundant—are crucial for modern molecular-genetic research, enabling applications such as GWAS, QTL mapping, and MAS. PACE, in particular, excels when analyzing a small number of SNPs across large sample sets, such as in sex determination, genetic mapping, and validation of crosses.
PCR Genotyping vs. Sequencing Methods
While next-generation sequencing (NGS) and genotyping-by-sequencing (GBS) are ideal for SNP discovery, PCR genotyping offers distinct advantages for routine marker validation and screening:
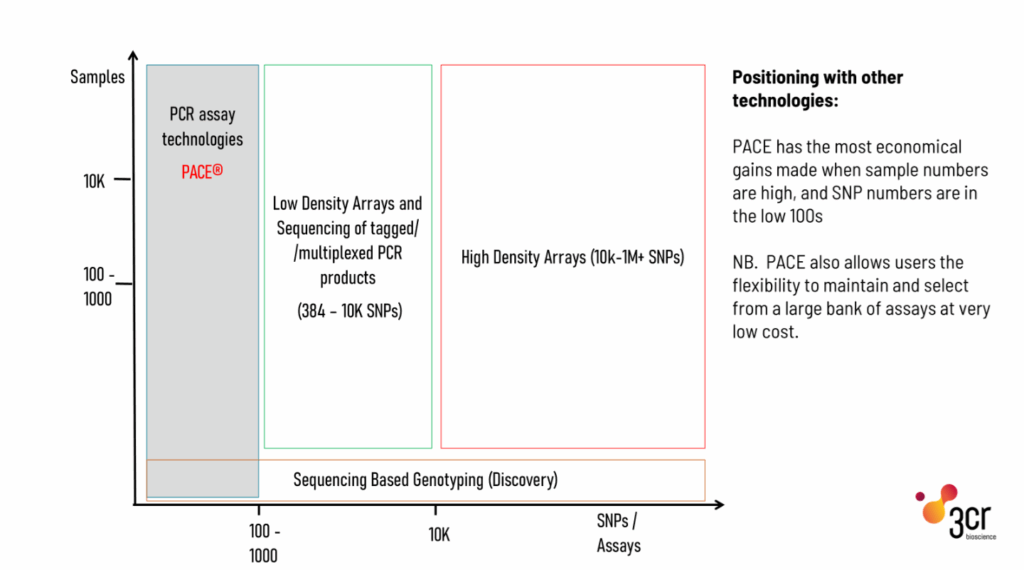
- Cost: PCR genotyping is significantly more affordable for moderate-scale projects (1-100 markers).
- Speed: Faster turnaround due to simplified analysis and minimal data processing.
- Data Volume: Sequencing generates large, often redundant datasets; PCR genotyping focuses on known, relevant SNPs.
- Scalability: PCR genotyping platforms like 3CR’s GeneArrayer efficiently processes hundreds to thousands of samples.
Applications of PCR Genotyping in Breeding Programs
In agriculture and livestock genetics, PCR-based SNP genotyping—especially using PACE—plays a pivotal role in:
- Marker Validation: Essential following SNP discovery via GBS or GWAS, confirming that markers are linked to traits of interest.
- Marker-Assisted Selection (MAS): Rapid screening for desirable traits, such as disease resistance, yield, or fertility.
- Genomic Selection (GS): Feeding validated SNPs into prediction models to estimate breeding values and drive genetic gain.
Advantages of PCR Genotyping with Allele-Specific PCR
- Precision and Specificity: Allele-specific PCR ensures accurate discrimination between alleles, critical for trait association.
- Flexibility: Custom primer design enables researchers to target specific loci relevant to breeding objectives.
- High Throughput: PCR genotyping can process large sample sizes efficiently without compromising on data quality.
- Cost-Effectiveness: Ideal for focused studies where sequencing is excessive or impractical.
Flexibility and Customization
Allele-specific PCR offers exceptional flexibility. Researchers can design primers targeting SNP loci associated with key breeding objectives, streamlining selection and improving outcomes. Its scalability supports diverse breeding projects—from targeted trials to population-wide screens.
As highlighted by von Maydell, primer design is a foundational step. A list of SNP-containing sequences can be submitted for rapid assay development. No reference genome is required, making PACE accessible for non-model organisms with limited genetic data.
Validation of Marker-Trait Associations
Accurate validation of SNP-trait links ensures the reliability of MAS strategies. Allele-specific PCR allows researchers to test associations across varied genotypes and environments. Including homozygous, heterozygous, and control samples, strengthens confidence in marker efficacy.
Integration with Genomic Selection
Validated SNP markers are key inputs in genomic prediction models. PCR genotyping provides the genotypic data needed to estimate breeding values, allowing earlier and more accurate selection. This synergy accelerates genetic improvement and enhances breeding efficiency.
SNP genotyping with allele-specific PCR is the preferred method for validating genetic markers in breeding programs, offering unmatched sensitivity, specificity, and customization options. By leveraging PCR genotyping, researchers and breeders can efficiently validate marker-trait associations, enhance genomic selection strategies, and expedite progress toward breeding objectives. In a field where efficiency and precision are crucial, adopting SNP genotyping with allele-specific PCR is a strategic investment in the future of agricultural and livestock breeding and genetic improvement.
Why Choose PACE® for PCR Genotyping?
PACE (PCR Allele Competitive Extension) from 3CR Bioscience represents a leap forward in PCR genotyping chemistry.
- Compatible with crude DNA samples
- Multiplexing capabilities
- Real-time signal observation
- One-step genotyping directly from RNA
- Interoperability with KASP™ and Amplifluor assays
PACE 2.0 offers enhanced signal clarity and tighter clustering especially with tricky or crude DNA samples, while PACE OneStep RT PCR enables simultaneous reverse transcription and genotyping in a single tube.
3CR Bioscience’s Genotyping Solutions:
- PACE Genotyping Master Mix: Universal mix with precise fluorescent signals across any reaction volume.
- PACE 2.0 Genotyping Master Mix: Improved signal-to-noise ratio and real-time reaction monitoring.
- PACE OneStep RT PCR Master Mix: Combines reverse transcription and PCR in one reaction for RNA-based genotyping.
- ProbeSure Master Mix: A cost-effective, robust hydrolysis probe mix for TaqMan™-style assays.
- Free assay design service: Rapid SNP/Indel marker development, with optional validation by our expert team.
Automating PCR Genotyping: 3CR’s GeneArrayer Platform
Automation significantly enhances PCR genotyping:
- Throughput & Speed: Thousands of samples processed daily with minimal hands-on time.
- Consistency: Standardized pipetting and thermal cycling enhance reproducibility.
- Scalability: Easily adaptable from small-scale validation to extensive population studies.
- Cost Efficiency: Reduced labour, waste, and reruns lower the per-sample cost.
3CR’s integrated platforms —GeneExtract, GeneArrayer, GeneCycler, and GeneScanner—enable seamless DNA processing from extraction to data analysis.
Case Studies: PACE in Action
Real-world applications of PACE PCR genotyping underscores its impact:
- Lamb Mortality Studies in Dairy Sheep: Early identification of genetic markers associated with survival rates.
- Boosting peanut cultivar development with PCR genotyping: Unlocking the full potential of peanuts with SNP genotyping and an allele-specific PCR workflow at Peanut Breeding & Genetics Program, Crop & Soil Sciences at, North Carolina State University
- Hop Breeding Sex Markers: PACE PCR genotyping boost hop breeding with an affordable and convenient diagnostic marker to identify male and female hop plants.
Conclusion: The Future of Genetic Analysis Lies in PCR Genotyping
PCR genotyping, particularly with PACE genotyping chemistry, bridges the gap between SNP discovery and trait selection, offering speed, affordability, and precision for life science researchers, biotech pioneers, agricultural researchers, and breeders alike. As von Maydell (2023) emphasizes, allele-specific systems like
PACE are ideal for reanalyzing validated SNPs across multiple genotypes—providing unmatched efficiency for genetic improvement.


