Polymerase Chain Reaction (PCR) is a cornerstone of molecular biology, enabling the amplification of specific DNA sequences. Understanding what a PCR master mix is can simplify your workflow, as this pre-formulated solution contains all the essential components needed for reliable and consistent PCR results.
What is a PCR Mastermix?
A PCR master mix simplifies molecular biology workflows by providing a convenient, all-in-one solution for PCR amplification. By including precisely measured ingredients such as DNA polymerase, nucleotides, buffer, and magnesium, PCR master mixes eliminate the need to prepare individual reagents, minimize human error, and improve the reproducibility of results. This makes PCR master mixes essential for both routine and high-throughput DNA analysis in research and clinical laboratories.
PCR Mastermix Ingredients
A standard PCR master mix typically includes:
- DNA Polymerase: The enzyme that synthesizes new DNA strands.
- dNTPs (Deoxynucleotide Triphosphates): The building blocks of DNA.
- MgCl₂ (Magnesium Chloride): An essential cofactor for polymerase activity.
- Buffer System: Maintains optimal pH and ionic conditions.
- Stabilizers and Enhancers: Enhance enzyme stability and reaction efficiency.
These components are optimized to ensure reliable PCR performance.
2x PCR Master Mix 4 Main Components
A 2x PCR master mix contains four main components essential for successful DNA amplification: DNA polymerase for copying DNA strands, deoxynucleotide triphosphates (dNTPs) as the building blocks of new DNA, buffer solution to maintain optimal reaction conditions, and magnesium ions (Mg²⁺) which are crucial for enzyme activity. This concentrated formulation allows you to simply add your template DNA and primers, streamlining the PCR setup while ensuring accuracy and consistency.ications.
2X PCR Master Mix Functions
A 2X PCR master mix is a concentrated version containing double the standard component concentrations. It allows users to add equal volumes of DNA template and primers, streamlining setup, and is ideal for high-throughput applications.
Choosing the Right PCR Master Mix
Selecting the best PCR master mix depends on your experimental goals, considering:
- Template Type: Genomic DNA, cDNA, or plasmid DNA.
- Application: Standard PCR, qPCR, or multiplex PCR.
- Sensitivity and Specificity: Some mixes are optimized for crude DNA preps, high sensitivity or fast cycling.
Why Use a Ready-to-Use PCR Master Mix?
- Convenience: No need for manual component preparation.
- Consistency: Reduces experimental variation.
- Optimized Performance: Formulated for robust amplification.
Discover High-Quality PCR Master Mixes at 3CR Bioscience
3CR Bioscience offers a range of optimized PACE PCR master mixes for diverse applications, ensuring highly accurate and reliable results.
What is PACE?
PACE® (PCR Allele Competitive Extension) is 3CR Bioscience’s patented genotyping chemistry specifically designed for Allele-Specific PCR genotyping. This technology is based on a Polymerase Chain Reaction (PCR) using two competing, un-labelled allele-specific primers, a common reverse primer, and an endpoint fluorescent measurement. PACE utilizes a novel, universal fluorescent reporting cassette in the PCR master mix to produce machine-readable fluorescent signals corresponding to genotypes, ensuring high accuracy in SNP and Indel detection. Unlike 5’ nuclease based probe-based products such as TaqMan™, PACE genotyping reactions only require un-labelled primer oligos to make up the PACE genotyping reaction, in combination with PACE Genotyping Master Mix, making them much more cost-effective, and quicker and easier to create new marker assays.
How PACE Chemistry Works
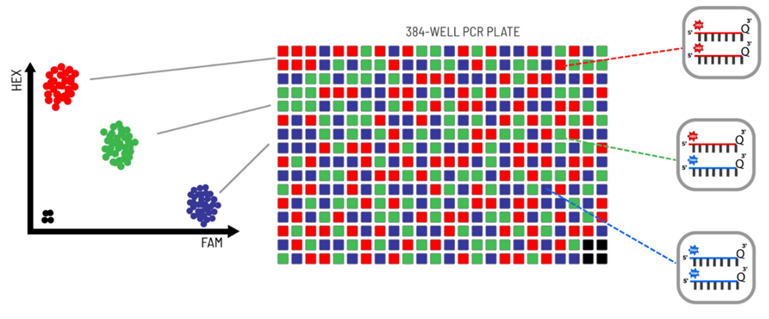
The PACE reaction starts with two allele-specific primers that compete to bind their 3’ ends at the SNP or Indel of interest. If the SNP is heterozygous, both primers can bind; if it is homozygous, only one primer binds. A common reverse primer binds to the opposite strand, ensuring complete amplification. As the PCR progresses, the fluorescent reporting cassettes in the PACE Genotyping Master Mix bind to the generated sequences, emitting signals specific to the alleles detected.
- Homozygous Genotype: A single fluorescent signal (FAM or HEX) is generated.
- Heterozygous Genotype: A combination of both fluorescent signals is produced.
Watch our quick video explaining How PACE genotyping works.


Components of a PACE Genotyping Reaction
A standard PACE genotyping reaction requires two main components:
- Custom PACE Genotyping Assay: Includes two allele-specific primers and a common reverse primer.
- PACE Genotyping Master Mix: A ready-to-use, optimized solution containing a specialized Taq polymerase, dNTPs, buffer, MgCl₂, and a universal fluorescent reporting cassette.
These components are combined with template DNA to enable efficient and accurate genotyping. The reaction can be analyzed using a qPCR machine or a fluorescent plate reader. PACE genotyping master mixes are designed for use with PACE Genotyping Assays and are also compatible with pre-existing KASP™ and Amplifluor® assays

Free PACE Assay Design Service
3CR Bioscience offers a free PACE Assay Design Service for customers who have purchased any PACE Master Mix within the last 24 months. By submitting your SNP/Indel of interest with flanking sequence, our scientific team will provide optimized assay designs tailored to your target sequences.
Unlike probe-based products such as TaqMan™, PACE genotyping reactions only require un-labelled primer oligos to make up the PACE genotyping reaction, in combination with PACE Genotyping Master Mix, making them much more cost-effective, and quicker and easier to create new marker assays.
If you are using 5’ nuclease and probe-based chemistry such as TaqMan, head over to our ProbeSure Paster Mix product page for the right mixes and information. ProbeSure™ Genotyping Master Mix is designed for use with 5’ nuclease assays and is suitable for use with TaqMan™, BHQ®, BHQplus®, Zen™ probes.
Explore our full range of PACE Genotyping Master Mixes
- PACE Genotyping Master Mix: A universal PCR master mix for allele-specific PCR assays. Precision fluorescent signal generation with consistently high performance at any reaction volume.
- PACE 2.0 Genotyping Master Mix: An enhanced PCR master mix for allele-specific assays. Improved signal to noise ratio and tight clustering. Developed specifically for genotyping direct from crude DNA samples.
- PACE Multiplex Master Mix: PACE Multiplex Master Mix for the simultaneous detection of up to four targets in one reaction. Save time, cost, and consumables while maximising data generation.
- PACE OneStep RT-PCR Master Mix: Genotype directly from RNA samples. RNA reverse transcription and cDNA PCR genotyping simultaneously in a single, one-step reaction.
- PACE Nano Master Mix: Advanced performance at ultra-low reaction volumes, delivering consistent, cost-effective PCR genotyping for high-throughput, miniaturised and automated workflows.
All our master mixes are supplied at 2x concentration for convenience and contain ROX normalising dye at either high (500 nM working concentration), low (25 nM working concentration), standard ROX level (150 nM working concentration) or without ROX. If a fluorescent plate reader is used instead of a qPCR instrument, it is recommended that the standard ROX version of the PACE Genotyping Master Mix is used.
You can find more information about which ROX level is right for your instrument in our ROX Instrument compatibility list here.
PACE Multiplex Master Mix
PACE Multiplex Master Mix is an advanced and versatile extension of our PACE 2.0 Genotyping Master Mix, formulated for the simultaneous detection of up to four targets per reaction, allowing for:
- Two biallelic SNPs in a single reaction.
- Three or four-allele SNPs.
- Three target genes plus a reference/housekeeping gene.
This multiplexing capability significantly increases throughput without compromising accuracy.
Users will require a plate reader capable of reading FAM, HEX, ATTO 590, ATTO 647N and reference dye ATTO 680 (wavelengths in the PACE Multiplex Master Mix User Guide). PACE Multiplex Master Mix is supplied at 2x concentration for convenience and with or without ATTO 680 reference dye at a range of levels to ensure compatibility with your qPCR machine or reader.
Practical Guide to Using PACE Master Mixes
1. Master Mix Storage and Shelf Life
- Store PACE master mixes at -20°C to -80°C for long-term stability.
- Avoid repeated freeze-thaw cycles to preserve enzyme activity.
- Protect the mix from light to maintain fluorescent signal integrity.
2. Arraying Template DNA
- Use a liquid-handling system or manually array DNA for low sample numbers.
- For high sample volumes, drying DNA can improve data consistency.
- Ensure complete drying for consistent results.
- A liquid handler can greatly improve speed, consistency and accuracy at low reaction volumes.
3. Assembling the PACE Genotyping Reaction
- Mix PACE Genotyping Master Mix, custom assays, and template DNA.
- Maintain a final 1x concentration of the master mix.
4. Dispensing and Sealing
- Dispense the reaction mixture into PCR plate wells.
- Seal plates with optically clear seals to prevent evaporation.
5. Thermal Cycling
- Follow the recommended thermal cycling protocol.
- For tighter clusters, add up to four additional cycles if needed.
6. Fluorescent Signal Detection
- Detect fluorescence with a qPCR machine or a fluorescent plate reader.
- Analyze using cluster analysis software or Microsoft Excel.
For full details and description of setting up and running genotyping reactions with PACE Genotyping Master Mix, please refer to the PACE Genotyping Master Mix User Guide.
Advanced Applications of PACE PCR Master Mixes.
Beyond SNP and Indel detection, PACE can be adapted for pathogen detection, transgenic sequence identification, and real-time monitoring. PACE master mixes are compatible with all major PCR genotyping platforms, including 96-, 384-, 1536- well PCR plates, as well as Array Tape®, producing accurate data regardless of the reaction volume.


